Solvated Fluorophore Dataset Notes¶
Solvents¶
| Solvent | \(\varepsilon_0\) | \(n\) | \(\alpha\) | \(\beta\) | \(\gamma\:(mN/m)\) \(293.15K\) |
\(\gamma\:(cal\cdot mol^{-1})\) \((mN/m\times 1.439)\) |
\(\phi\) | \(\psi\) | Toby owns | Needs purchasing | Price |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N-Hexane | 1.88 | 1.3749 | 0.00 | 0.00 | 18.35 | 26.41 | 0.00 | 0.00 | |||
| Toluene | 2.37 | 1.4969 | 0.00 | 0.14 | 27.94 | 40.21 | 0.86 | 0.00 | |||
| Anisole | 4.22 | 1.517 | 0.00 | 0.29 | 35.37 | 50.90 | 0.75 | 0.00 | |||
| Diethyl ether | 4.24 | 1.3473 | 0.00 | 0.41 | 17.15 | 24.68 | 0.00 | 0.00 | |||
| \(\ce{CH3Cl}\) | 4.71 | 1.4458 | 0.15 | 0.02 | 27.10 | 39.00 | 0.00 | 0.50 | |||
| THF | 7.43 | 1.4072 | 0.00 | 0.48 | 27.37 | 39.39 | 0.00 | 0.00 | $150-240/L | ||
| DCM | 8.93 | 1.4241 | 0.10 | 0.05 | 27.84 | 40.06 | 0.00 | 0.67 | |||
| Ocanol | 9.86 | 1.430 | 0.37 | 0.48 | 26.02 | 37.44 | 0.00 | 0.00 | $100-190/L | ||
| EtOH | 24.85 | 1.3614 | 0.37 | 0.48 | 22.28 | 32.06 | 0.00 | 0.00 | |||
| ACN | 35.69 | 1.3441 | 0.07 | 0.32 | 28.37 | 40.82 | 0.00 | 0.00 | |||
| DMF | 37.22 | 1.4305 | 0.00 | 0.74 | 36.73 | 52.85 | 0.00 | 0.00 | |||
| DMSO | 46.83 | 1.4793 | 0.00 | 0.88 | 43.36 | 62.40 | 0.00 | 0.00 |
There’s no point in doing water, since SMD uses its own parameters for it and apart from nile red, nothing will fluoresce in it.
Should we use octanol? I wanted a low dialectric, h-bonding solvent, but maybe Michelle’s comments suggest not to add more h-bonding solvents.
The Initial Proposed dataset¶
| Fluorophore | Toby owns | Needs digging up | Needs purchasing | Class | Other notes | Price |
|---|---|---|---|---|---|---|
| Rhodamine 6G | Rhodamine | |||||
| Rhodamine 123(?) | Rhodamine | |||||
| AlexaFluor 532 | Rhodamine-like | In azide form | ||||
| NDI (of some description) | NDI | |||||
| Naphthalamide (of some description) | NDA | |||||
| Prodan/ | Prodan | Prodan $245/25mg | ||||
| DAPI | DAPI | “Someone in bio will have some” | $128/10mg | |||
| FITC | Fluorescein | Can we just use fluorescein? | $66 USD/100mg | |||
| Coumarin 343/519 | Coumarin | |||||
| Texas Red | Rhodamine-like | “Alison will have some” | $256/5mg | |||
| Nile Red | Oxazine | |||||
| BODIPY 493/503 | BODIPY | Toby wants this to be investigated | $467/500mg | |||
| Merocyanine 540 | Cyanine | |||||
| Dansyl | Naphthalene | Which dansyl? | Hydrazine $68/250mg Amide $150/1g Chloride $160/1g |
|||
| Azulene | Azulene | \(S_2\to S_0\) emitter | $55/50mg | |||
| Indigo Carmine | $87/25g | |||||
| Cascade Blue | Pyrene | $208/1g |
After Consultation with Michelle¶
Notes form meeting
Suggestions to the dataset¶
- Add quinones and anthroquinones,
- Add aza-indole
- Remove merocynainie
- remove anything that can internally hydrogen bond and any solvents that will hydrogen bond
- We Don’t want s1-s0 gap being too small - biradicals
- Find species in the dataset that enolise - we don’t really want this
- make sure there’s a few ct molecules, but not too many * [ ] Probably want a subset of CT molecules
- Need to consider how much of the emisison state is populated
Suggestions for the process¶
- Cluster continuum will remove some of the issues with h-bonding - “That would be more satisfactory”
- For a benchmarking referecne, photoelectron spectra in gas phase to determine the \(t_1 \to s_0\) gap
- For casscf - basis set def2-tzvp
- For dft - use cam-b3lyp - not too small of a basis set * Need to consider more roots. the errors are going to be much worse in higher roots. * This brings a bigger question - should we be comparing the WHOLE spectra, instead of just the lower states/0-0 transition * Probably need caspt2 results with casscf geoms
To read up on¶
- Papers by jackerman
-
El-sayad rules
-
The rate of intersystem crossing is relatively large if the radiationless transition involves a change of orbital type
- e.g. \(^1\pi,\pi^* \to ^3n,\pi^*\) is faster than \(^1\pi,\pi^* \to ^3\pi,\pi^*\)
-
- Danny jacoman’s work benchmarking dft
- Marco garavelli’s work
To investigate¶
- Check the difference between wb97x-dx and wb97x
If I had a system…
- I’d look at the structure and look and look at the solvent, and specifically use cluster continuum.
- I’d look at molecules that don’t internally hydrogen bond
- I’d use the best method I could, and if that’s not MR, I’d use a selection of DFT functionals
- I’d then try to find the combination of method and solvation method that gives the best results.
We have regularly come to the conclusion that SMD does a good job and that wb97-xd does a decent job - if there’s no hydrogen bonding involved. ” I don’t believe for a moment in accurate quantitative”
The Revision Process:¶
| Fluorophore | Told to remove | Internal/H-Bonding acceptor/donor | Protonation sites | \(pK_a\) | (un)Cyclises | Do I want it |
|---|---|---|---|---|---|---|
| Rhodamine 6g | They seem to be non-fluorescent in their closed form though… | |||||
| Rhodamine 123 | ||||||
| Rhodamine 800 | ? | |||||
| AlexaFluor 532 | ||||||
| NDI | ||||||
| Naphthalamide | ||||||
| Prodan | 2? | |||||
| DAPI | ? | 4.31 | ||||
| Coumarin 343/519 | 1-3 | |||||
| 7-amino-4-methylcoumarin (coumarin 120) | 1-2 | 3.37 | ||||
| Texas Red | many | many | ||||
| Nile Red | 1 | 4.08 | ||||
| BODIPY 493/503 | ||||||
| Dansyl Amide | 4.63/9.97 | |||||
| Azulene | 0 | |||||
| Indigo Carmine | 2 | 12-13 | ||||
| Fluorescein | ||||||
| FITC | ||||||
| Merocyanine | ||||||
| Cascade Blue | ||||||
| 1-aminoanthroquinone | 1 | |||||
| aza-indole | 4.59 | |||||
| anthracene? | 0 |
Rejected¶
| Fluorophore | Toby owns | Needs digging up | Needs purchasing | CT | Class | Other notes | Price |
|---|---|---|---|---|---|---|---|
| 7-azaindole | indole | $47.80/1g |
New Dataset¶
| Fluorophore | Toby owns | Needs digging up | Needs purchasing | CT | Class | Other notes | Price |
|---|---|---|---|---|---|---|---|
| Rhodamine 800 | Rhodamine | $216/250mg | |||||
| NDI (of some description) | NDI | ||||||
| Naphthalamide (of some description) | NDA | ||||||
| Prodan | Prodan | $245/25mg | |||||
| DAPI | DAPI | “Someone in bio will have some” | $128/10mg | ||||
| Coumarin 153 | Coumarin | ||||||
| Nile Red | Oxazine | ||||||
| BODIPY 493/503 | BODIPY | Toby wants this to be investigated | $467/500mg | ||||
| Dansyl Amide | Naphthalene | $150/1g | |||||
| Azulene | Azulene | \(S_2\to S_0\) emitter | $55/50mg | ||||
| Indigo Carmine | $87/25g | ||||||
| 1-aminoantrhaquinone | anthraquinone | $76.30/100g |
Other option¶
| Fluorophore | Toby owns | Needs digging up | Needs purchasing | Class | Other notes | Price |
|---|---|---|---|---|---|---|
| Anthracene | anthracene | Can see the fine structure, only soluble in low-polar solvents | $42.2/1g |
Fluorophore Specific Notes¶
Notes of concern¶
- Rhodamine 800
- Concentrations need to be kept low to prevent aggregation
- Seems to be poorly handled by TD-DFT, but I think this should be okay
- The \(\ce{ClO4-}\) might have some chemical interactions that should lead us to exclude this species.
- 7-Azaindole
- Forms clusters in solution and tautomerises with any protic solvent facilitate it
- BODIPY - all varieties and cyanine dyes (N-B-N, N-B-O and O-B-O)
- Have strong MR character from double-excitation that leaved the adiabaitc TD-DFT approach with large errors 0.3-0.5 eV
Finalised!¶
| Fluorophore | Solvated Dataset | Gas Dataset | Class | \(m\)-diagnostic | \(D_{CT}\:(\AA)\) | \(t\:(\AA)\) |
|---|---|---|---|---|---|---|
| Azulene | Yes | Yes | Azulene | 0.120 | 0.967 | -0.859 |
| Rhodamine 800 | Yes | No | Xanthene | 0.076 | 1.074 | -0.436 |
| 1-aminoanthraquinone | Yes | No | Quinone | 0.063 | 2.914 | 1.377 |
| Coumarin 153 | Yes | Yes | Coumarin | 0.087 | 1.877 | -0.271 |
| Nile Red | Yes | No | Oxazine | 0.078 | 1.592 | -1.376 |
| BODIPY 493/503 | Yes | No | Cyanine | 0.087 | 0.453 | -0.941 |
| N-propyl-4-hydroxyl-1,8-naphthalamide | Yes | No | Naphthalamide | 0.071 | 0.626 | -1.197 |
| DAPI | Yes | No | Phenylindole | 0.082 | 1.959 | -1.249 |
| Dansyl Amide | Yes | No | Dansyl | 0.071 | 1.660 | -0.087 |
| Boron subphthalocyanine chloride | Yes | No | Cyanine | 0.096 | 0.398 | -1.830 |
| α-Sexithiophene | Yes | No | Polymeric thiophene | 0.079 | 0.002 | -5.353 |
| Rhodamine 575 | No | Yes | Xanthene | 0.050 | 0.741 | -1.228 |
| Fluorescein | No | Yes | Xanthene | 0.074 | 3.756 | 2.358 |
| 8-methoxy-BODIPY | No | Yes | Cyanine | 0.089 | 0.770 | -1.209 |
| Fluorophore | Price | Quantity | Shop | Item Number | Notes | Recieved |
|---|---|---|---|---|---|---|
| Already have | ||||||
| Nile Red | ||||||
| Naphthalamide | ||||||
| Rhodamine 800 | $170.10 | 250mg | Sigma | 8370 | ||
| Coumarin 153 | $57.96 | 100mg | Sigma | 546186 | ||
| BODIPY 503 | $368.10 | 500mg | Sigma | 790389 | ||
| Azulene | $44.01 | 50mg | Sigma | 37879 | ||
| 1-aminoanthraquinone | $60.21 | 100g | Sigma | A39009 | ||
| DAPI | $287.13 | 10mg | ThermoFisher | 62247 | ||
| Dansyl Amide | $117.90 | 1g | Sigma | 218898 | ||
| Boron subphthalocyanine chloride | $104.03 | 50mg | Biosynth | FB179000 | ||
| α-Sexithiophene | $26 USD | 100mg | TCI | S0504 | ? | |
| Total | ~$1248.31 AUD |
Rejected species Types¶
- Fluoresceins - internal cyclisation - require pH modification
Rejected species¶
- 7-azaindole
- indigo species - Tautomerise freely and have a very high triplet yield
- It’s always depicted with the perchlorate ion, but I’m not sure if that’s significant for its fluorescence, or if it’s just the common counterion.
DOI: 10.1016/j.jlumin.2012.08.017
- Has a relatively small stokes shift, with mostly two identifiable emission peaks though the tail extends all the way to 950 nm
- Is VERY red
- Φ of ~0.25 in ethanol
DOI: 10.1007/s11664-018-6367-6
- Has a pretty small solvatochromic effect
- Concentration dependent, with 23nm redshifting ocurring as aggregates form (mM or greater)
DOI: 10.1002/qua.25780
“Why the lowest electronic excitations of rhodamines are overestimated by time-dependent density functional theory”
- They seem to have large double excitation character, even in their singlet states, so MR and highly correlated methods can capture this, but TD-DFT struggles a bit.
- Has a “partial” CT character? (0.37e over 1.791Å)
- Not entirely sure how much I trust all of this without using cLR/VEM though.
- They used modest basis sets for their testing 6-31+G(d,p)
- Haven’t specified integration grids, which makes me think they used the G09 default (pruned 75,302) which probably isn;t sufficient for all their functionals
- Used the same B3LYP-D3 geom for all their functionals
DOI: 10.1007/s00894-022-05034-w
- TD-DFT paper in water. B3PW91/6-31++G(d,p)/IEFPCM
- Has an H-bonding site on the nitrile that needs attention
DOI: 10.1021/ct500775r DOI: 10.1021/ar500447q
- In general, these seem to have double-excitation character in the lowest ES, making the adiabatic approximation of TD-DFT a big ask.
- Not CT species
- Very large Φ
-
Looks like have a large MR character
- Need either correlated or MR methods to model
- Might not be the most appropriate for a TD-DFT datadset
REALLY useful thesis on Azulene
DOI: 10.1016/0009-2614(74)85131-6
- Has mostly single fluorescence (\(s_2 \to s_0\)) unless substituted
- Energy gap between \(s_2\) and \(s_1\) is large enough that \(s_2 \to s_2\) non-radiative relaxation is too slow to compete with \(s_2 \to s_0\) fluorescence
- Conical intersection between \(s_1\) and \(s_0\)
- Small amounts of \(S_1\) emission can be detected as \(t_1 \to s_1 \to s_0\)
- Both absorbance and emission are highly solvatochromic - large Stokes shifts
- Φ decreases with increasing solvent polarity (0.15 in n-hexane to <0.03 in DMSO)
- Charge Transfer species
- Amine can either be twisted (TICT) or planar (PICT) - low energy barrier
- Excited state intramolecular proton transfer not considered possible in \(s_1\)
Timing
- 560 ps fluorescence lifetime
- 4.7 ps vibrtational relaxation in the ICT state
- 150-180 ps ICT dynamics
- CAM-B3LYP/6-31G(d,p) (no diffuse functions?)
- LR IEF-PCM ACN
Results
- Identified the \(s_1\) state as the “crucial state”, as it’s the lowest optically allowed excited state with a “relatively large oscilaltor strength”
- In the \(s_1\) state, it looks like the amino group wants to be perpendicular to the rings of the molecule
- In polar solvents, it looks like they quench emission by enhancing ISC from \(s_1 \to t_2\). I’m guessing this is die to a competition between intramolecular H-bonding and solvent-solute H-bonding
<iframe style="width: 100%; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=9222&bg=white"></iframe>
DOI: [10.1021/jp9630232](https://doi.org/10.1021/jp9630232)
* Forms clusters in solution with water and alcohols that have different $\lambda_{max}^{em}$
* Water will pronate this species... not sure if other solvents can do this, as it seems to be largely cluster-shape dependent
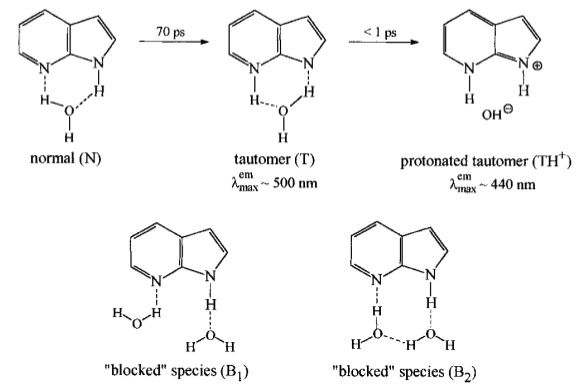{class="center"}
Timing
* 910 ps fluorescence lifetime in water at pH=7
DOI: [10.1021/j100178a023](https://doi.org/10.1021/j100178a023)
* It also seemingly tautomerises when excited, so I think we should rule out this species unless we go for aprotic only solvents